Researchers at MIT have introduced a polymer that stores DNA at room temperature, bypassing the need for costly freezing. This development supports sustainable long-term storage and damage-free retrieval of both genetic material and digital data. Credit: SciTechDaily.com
With their “T-REX” method, 
The glassy, amber-like polymer can be used for long-term storage of DNA, such as entire human genomes or digital files such as photos. Credit: MIT News; iStock
Streamlining DNA Preservation Techniques
“Freezing DNA is the number one way to preserve it, but it’s very expensive, and it’s not scalable,” says James Banal, a former MIT postdoc. “I think our new preservation method is going to be a technology that may drive the future of storing digital information on DNA.”
Banal and Jeremiah Johnson, the A. Thomas Geurtin Professor of Chemistry at MIT, are the senior authors of the study, published on June 12 in the Journal of the American Chemical Society. Former MIT postdoc Elizabeth Prince and MIT postdoc Ho Fung Cheng are the lead authors of the paper.
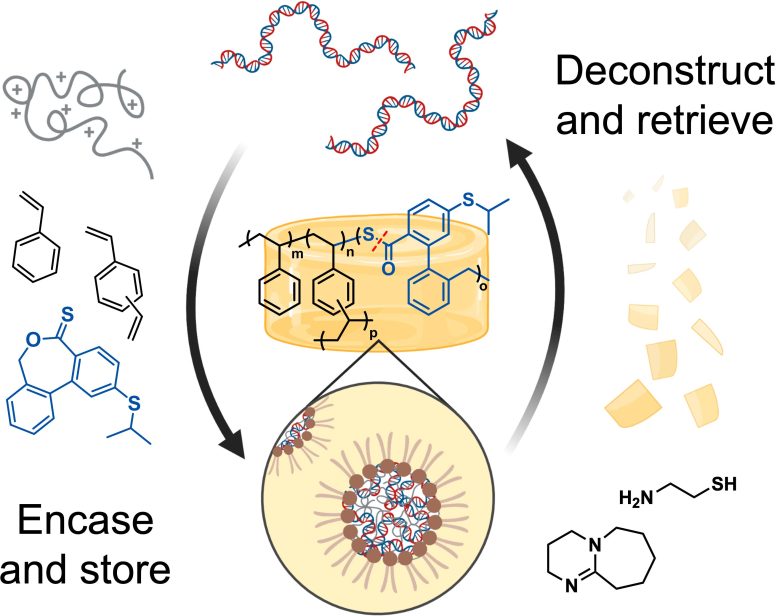
MIT researchers have devised a way to encapsulate DNA into a thermoset polymer known as cross-linked polystyrene. After the DNA is embedded into the polymer, it can be released again by treating the polymer with cysteamine. Credit: Courtesy of the researchers
Exploring New DNA Encoding Methods
DNA, a very stable molecule, is well-suited for storing massive amounts of information, including digital data. Digital storage systems encode text, photos, and other kind of information as a series of 0s and 1s. This same information can be encoded in DNA using the four nucleotides that make up the genetic code: A, T, G, and C. For example, G and C could be used to represent 0 while A and T represent 1.
DNA offers a way to store this digital information at very high density: In theory, a coffee mug full of DNA could store all of the world’s data. DNA is also very stable and relatively easy to synthesize and sequence.
In 2021, Banal and his postdoc advisor, Mark Bathe, an MIT professor of biological engineering, developed a way to store DNA in particles of silica, which could be labeled with tags that revealed the particles’ contents. That work led to a spinout called Cache DNA.
One downside to that storage system is that it takes several days to embed DNA into the silica particles. Furthermore, removing the DNA from the particles requires hydrofluoric DOI: 10.1021/jacs.4c01925
The research was funded by the National Science Foundation.













/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/filer_public/d1/82/d18228f6-d319-4525-bb18-78b829f0791f/mammalevolution_web.jpg)






Discussion about this post